|
read_par
This routine reads the `NSIDE', `TFIELDS' , `MAX-LPOL', and optionally `MAX-MPOL'
keywords from a FITS-file. These keywords desribe Nside, number of
datasets (maps) and maximum multipole l (order) and m (degree) value
for the file. If a keyword is not found in the FITS file, a value of -1 is
returned instead. The file could eg. be a HEALPix map, or a set of alm or precomputed
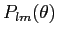
Location in HEALPix directory tree:
src/f90/mod/fitstools.f90
FORMAT call read_par(
filename, nside, lmax, tfields [, mmax]
)
ARGUMENTS
| name & dimensionality |
kind |
in/out |
description |
|---|
| |
|
|
|
|
filename(LEN=filenamelen) |
CHR |
IN |
filename of the FITS file. |
| nside |
I4B |
OUT |
`NSIDE' keyword value from the FITS header. |
| lmax |
I4B |
OUT |
`MAX-LPOL' keyword value from the FITS header. |
| tfields |
I4B |
OUT |
`TFIELDS' keyword value from the FITS header. |
| mmax (OPTIONAL) |
I4B |
OUT |
`MAX-MPOL' keyword value from the FITS header. |
EXAMPLE:
call read_par('plm_128p.fits', nside, lmax, nhar)
Checks the Nside and maximum l value used for the precomputed
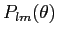 that are stored in the file `plm_128p.fits'. If the file also contains tensor harmonics, nhar is returned with the value 3, otherwise it is 1. that are stored in the file `plm_128p.fits'. If the file also contains tensor harmonics, nhar is returned with the value 3, otherwise it is 1.
MODULES & ROUTINESThis section lists the modules and routines used by read_par.
-
fitstools
- module, containing:
-
printerror
- routine for printing FITS error messages.
-
cfitsio
- library for FITS file handling.
RELATED ROUTINESThis section lists the routines related to read_par
-
synfast, plmgen
- executables that produce FITS-files with keywords relevant to this routine.
Eric Hivon
2010-06-18
|

|