|
read_tqu
This IDL facility reads a temperature+polarization Healpix map
(T,Q,U) from a binary table FITS file,
with optionally the error (dT,dQ,dU) and correlation (dQU, dTU, dTQ)
from separate extensions
Location in HEALPix directory tree:
src/idl/fits/read_tqu.pro
FORMAT IDL>
READ_TQU , File, TQU, [Extension=, Hdr=, Xhdr=, Help=, Nside=, Ordering=, Coordsys=]
QUALIFIERS
-
File
- name of a FITS file from which the maps are to be read
-
TQU
- : array of Healpix maps of size (
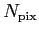 ,3,n_ext) where ,3,n_ext) where 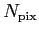 is the total
number of Healpix pixels on the sky, and n_ext is the total
number of Healpix pixels on the sky, and n_ext 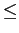 3 is
the number of extensions read 3 is
the number of extensions read
Three maps are available in each extension of the FITS file :
-the temperature+polarization Stokes parameters maps (T,Q,U) in
extension 0
-the error maps (dT,dQ,dU) in extension 1 (if applicable)
-the correlation maps (dQU, dTU, dTQ) in extension 2 (if applicable)
-
Extension=
- (optional),
extension unit from which to read the data (0 based).
If absent, all available extensions are read
-
Hdr=
- (optional),
string variable containing on output the contents of the primary header. (If already present, FITS reserved
keywords will be automatically updated).
-
Xhdr=
- (optional),
string variable containing on output the contents of the
extension header. If
several extensions are read, then the extension
headers are returned appended into one string array.
-
Nside=
- (optional),
returns on output the HEALPix resolution parameter, as read
from the FITS header. Set to -1 if not found
-
Ordering=
- (optional),
returns on output the pixel ordering, as read from the FITS
header. Either 'RING' or 'NESTED' or ' ' (if not found).
-
Coordsys=
- (optional),
returns on output the astrophysical coordinate system used,
as read from FITS header (value of keywords COORDSYS or SKYCOORD)
KEYWORDS
-
Help
- if set, an extensive help is displayed and no
file is read
DESCRIPTION
read_tqu reads out Stokes parameters (T,Q,U) maps for the whole
sky into a FITS file. It is also possible to read the error per pixel for each
map and the correlation between fields, as subsequent extensions of the same FITS
file (see qualifiers above). Therefore the file may have up to three extensions with three
maps in each. Extensions can be written together or one by one (in
their physical order) using the Extension option
RELATED ROUTINESThis section lists the routines related to read_tqu
-
idl
- version 6.1 or more is necessary to run read_tqu
-
synfast
- This HEALPix f90 facility can be used to generate
temperature+polarization maps that can be read with read_tqu
-
write_tqu
- This HEALPix IDL facility can be used to write
out temperature+polarization that can be read by read_tqu.
-
read_fits_cut4,
read_fits_map
-
read_tqu,
read_fits_s
- HEALPix IDL routines to read cut-sky maps, full-sky maps, polarized full-sky maps and
arbitrary data sets from FITS files
-
read_fits_s
- This general purpose HEALPix IDL facility can be used to read
into an IDL structure maps contained in binary table FITS files.
-
sxpar
- This IDL routine (included in HEALPix package) can be
used to extract FITS keywords from the header(s) HDR or XHDR read with read_tqu.
EXAMPLE:
Reads into TQU the polarization maps contained in the FITS file
'map_polarization.fits'.
The variable xhdr will contain the extension(s) header.
Eric Hivon
2010-06-18
|

|