
|
anafast
This program performs harmonic analysis of the HEALPix maps
up to a user
specified maximum spherical harmonic order lmax.
The integrals are computed on the whole sphere, unless the user
chooses a provided option
to excise from the input map(s) a simple, constant latitude, symmetric cut, and/or
apply an arbitray cut read from an external file.
Scalar, or scalar and tensor, spherical harmonic coefficients are evaluated
from the map(s) if the input provides, respectively, only the temperature,
or temperature and polarisation maps.
The total operation count scales as
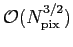 with a prefactor
depending on lmax. with a prefactor
depending on lmax.
Anafast reads one (two) file(s) containing the map(s) and produces
a file containing the temperature auto- (or cross-) power spectrum
CTl and, if requested,
also the polarisation power spectra CEl, CBl,
CT x El,
CT x Bl and
CE x Bl.
The alm coefficients computed during the execution also can be
written to a (two) file(s) if requested.
Anafast executes an approximate, discrete point-set quadrature on
a sphere
sampled at the HEALPix pixel centers.
Spherical harmonic transforms are computed
using recurrence relations for Legendre polynomials on co-latitude,
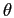 ,
and Fast Fourier Transforms on longitude, ,
and Fast Fourier Transforms on longitude, 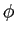 . .
Anafast is provided with an option to use precomputed Legendre Polynomials
to decrease the total execution time.
Anafast permits two execution options
which allow a significant improvement of accuracy of
the approximate quadrature performed by this facility:
 An improved analysis using the provided ring weights,
which correct the quadrature
on latitude, and/or An improved analysis using the provided ring weights,
which correct the quadrature
on latitude, and/or
 An iterative scheme using in
succession several backward and forward harmonic transforms
of the maps. An iterative scheme using in
succession several backward and forward harmonic transforms
of the maps.
Location in HEALPix directory tree:
src/f90/anafast/anafast.f90
RECOMMENDATIONS FOR USERS
Execution of anafast requires a user to specify the maximum
spherical harmonic order lmax up to which the harmonic
decomposition of the input maps will be performed.
Since there are no formal limits on parameter
lmax enforced by anafast, the user should make his/her choices
judiciously.
Hereafter it is convenient to specify lmax
in terms of the HEALPix
map resolution parameter nsmax.
If the function to be analysed is strictly band-width
limited, or nearly band-width limited (as in the case of
a Gaussian beam smoothed signal discretized at a rate of a few pixels
per beam area), it is sufficient to run anafast with
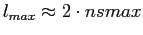 , with a very good Cl
error performance
already in the raw (i.e. uncorrected quadrature) harmonic transform mode.
If quadrature
corrections are still desired in this case, it should be sufficient to use, at no
extra cost in execution time, the ring-weighted quadrature scheme.
This is the recommended mode of operation of anafast for essentially
error and worry free typical applications, e.g. CPU-intensive
Monte Carlo studies. , with a very good Cl
error performance
already in the raw (i.e. uncorrected quadrature) harmonic transform mode.
If quadrature
corrections are still desired in this case, it should be sufficient to use, at no
extra cost in execution time, the ring-weighted quadrature scheme.
This is the recommended mode of operation of anafast for essentially
error and worry free typical applications, e.g. CPU-intensive
Monte Carlo studies.
If more aggressive attempts are undertaken to extract from a map
the spectral coefficients at
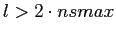 (for example, as
in a possible case of an attempt to analyse an existing map, which was
irreversibly binned at a suboptimal resolution)
the following should be kept in mind: (for example, as
in a possible case of an attempt to analyse an existing map, which was
irreversibly binned at a suboptimal resolution)
the following should be kept in mind:
 Spherical harmonics discretized using HEALPix
(either sampled at
pixel centers, or avaraged over pixel areas) form a linearly independent
system up to Spherical harmonics discretized using HEALPix
(either sampled at
pixel centers, or avaraged over pixel areas) form a linearly independent
system up to
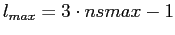 . Hence, the functions which are
strictly band-width limited to . Hence, the functions which are
strictly band-width limited to
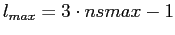 can be fully
spectrally resolved with anafast, albeit with integration errors
in the uncorrected quadrature mode, which grow up to
can be fully
spectrally resolved with anafast, albeit with integration errors
in the uncorrected quadrature mode, which grow up to
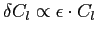 , with , with 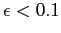 ,
at the highest values of l. These integration errors
can be efficiently
reduced
using anafast in the iterative mode. Although this lmax range
-- ,
at the highest values of l. These integration errors
can be efficiently
reduced
using anafast in the iterative mode. Although this lmax range
--
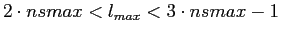 -- is easily
manageable with anafast used on strictly band-width limited functions,
it should be used with caution in basic and automated applications, e.g.
Monte Carlo simulations. -- is easily
manageable with anafast used on strictly band-width limited functions,
it should be used with caution in basic and automated applications, e.g.
Monte Carlo simulations.
 As with any discrete Fourier transform, anafast application to
functions which are not band-width limited results with aliasing
of power, which can not be remedied. If the particular case of interest
may result in such a band-width violation (i.e. there is significant power
in the function at As with any discrete Fourier transform, anafast application to
functions which are not band-width limited results with aliasing
of power, which can not be remedied. If the particular case of interest
may result in such a band-width violation (i.e. there is significant power
in the function at
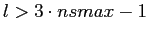 ), the function should
be smoothed before the application of anafast, or discretized and
then analysed, on a refined HEALPix grid (with larger nsmax). ), the function should
be smoothed before the application of anafast, or discretized and
then analysed, on a refined HEALPix grid (with larger nsmax).
 REMEMBER:
A peculiar property of the sphere, which usually surprizes those
whose intuition is built on experience with FFTs on a segment, or
on a Euclidean multidimensional domains, is the lack of
a regular and uniform point-set at arbitrary resolution,
and the resulting non-commutativity of the forward and
backward discrete Fourier transforms on nearly-uniform point-sets,
e.g. HEALPix . Hence,
as in any case of attempting an extreme application of an off-the-shelf
software, use caution and understand your problem well before
executing anafast under such cicumstances! REMEMBER:
A peculiar property of the sphere, which usually surprizes those
whose intuition is built on experience with FFTs on a segment, or
on a Euclidean multidimensional domains, is the lack of
a regular and uniform point-set at arbitrary resolution,
and the resulting non-commutativity of the forward and
backward discrete Fourier transforms on nearly-uniform point-sets,
e.g. HEALPix . Hence,
as in any case of attempting an extreme application of an off-the-shelf
software, use caution and understand your problem well before
executing anafast under such cicumstances!
FORMAT %
anafast [options] [parameter_file]
COMMAND LINE OPTIONS
-
-d
-
--double
- double precision mode (see Notes on double/single precision modes on page
![[*]](crossref.png) ) )
-
-s
-
--single
- single precision mode (default)
QUALIFIERS
-
infile =
- Defines the input map file.
(default= map.fits)
-
infile2 =
- Defines the 2nd input map file, to be cross-correlated with
the first one. The 2 maps should match in resolution (nsmax) and coordinate.
(default= `', only the auto-correlation of the first map will be computed)
-
outfile =
- Defines the output file with the power spectrum. If only
one input map is provided, outfile will contains its auto-spectra,
if 2 maps are provided, outfile will contain their
cross-spectra. Note in particular that in the latter case, the
CT x El power
spectrum will be build from the T field of the 1st (possibly polarized) map, and the E
field of the second polarized map.
(default= cl_out.fits)
-
simul_type =
- Defines which map(s) to analyse, 1=temperature only, 2=temperature AND polarisation.
(default= 1)
-
nlmax =
- Defines the maximum l value
to be used. See the Recommendations for Users.
(default= 64)
-
maskfile =
- Defines the FITS file containing the pixel mask(s) or
weighting scheme(s) by which the map(s) read from infile will be
multiplied before analysis. If the file contains several fields, the first
one in which at least one pixel is non-zero will be used. This option can be
used to, for instance, apply
the WMAP Kp intensity mask to the data (see
http://lambda.gsfc.nasa.gov
), but it will not handle the WMAP composite mask correctly.
Can be used in conjonction with theta_cut_deg. Masked or weighted pixels
will be correctly accounted when performing the monopole and dipole regression.
(default= `': no mask, all valid pixels are used)
-
theta_cut_deg =
- Defines the latitude (in degrees) of
an optional, straight symmetric cut around the equator. Pixels located within
that cut (|b|<theta_cut_deg are ignored.
(default=
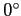 : no cut) : no cut)
-
iter_order =
- Defines the maximum order of quadrature
iteration to be used. (default=0, no iteration)
-
outfile_alms =
- Defines the name of the file
containing the alm coefficients of the first map
which can be written optionally. (default= no entry --
alms are not written to a file)
-
outfile_alms2 =
- Defines the name of the file
containing the alm coefficients of the second map, if any,
which can be written optionally. (default= no entry --
alms are not written to a file)
-
plmfile =
- Defines the name for an input file
containing precomputed Legendre polynomials Plm.
(default= no entry -- anafast executes the recursive evaluation
of Plms)
-
w8file =
- Defines name for an input file containing ring
weights in the improved quadrature mode (default= no entry --
the name is assumed to be 'weight_ring_n0xxxx.fits' where xxxx is nsmax)
-
w8filedir =
- Gives the directory where the ring weight files are
to be found (default= no entry -- anafast searches the default
directories, see introduction)
-
won =
- Set this to 1 if ring weight files are to be used,
otherwise set it to 0 (or 2). (default= 0)
-
regression =
- Sets the degree of the regression made on the
input map before doing the power spectrum analysis.
The regression is a minimal variance fit (assuming a uniform noise)
made on valid (unflagged and unmasked) pixels, out of the symmetric cut (if
any). In case of cut sky analysis, such a regression reduces the monopole
and dipole leakage to higher l's.
0 : no regression, does the a_lm analysis on the raw map
1 : removes the best fit monopole first
2 : removes the best fit monopole and dipole first
default = 0.
DESCRIPTION
Anafast reads one of two binary FITS-files containing a HEALPix map. These
files can each contain
a temperature map or both temperature and polarisation (Q,U) maps. Anafast analyses
the map(s) and makes an output ascii-FITS file containing the angular auto or cross
power spectra CTls
(and CEl, CBl,
CT x El,
CT x Bl and
CE x Bl if specified). Anafast
produces Cls up to a specified maximum l-value
(see Recommendations for Users). Note in particular that if 2 maps are provided,
the
CT x El power
spectrum will be build from the T field of the 1st (polarized) map, and the E
field of the second polarized map.
If requested, the computed alm coefficients
can be written to a FITS file. This file can be used in the
constrained realisation mode of synfast.
Anafast permits two execution modes that allow to improve
the quadrature accuracy:
(1) the ring weight corrected quadrature, and
(2) the iterative scheme.
Using the ring weights does not increase the execution time.
The precomputed ring weights to be used for each
HEALPix resolution nsmax are provided in
the $HEALPIX/data directory.
The more sophisticated iterative scheme increases the
accuracy more effectively than the weighted ring scheme,
but its disadvantage is that the time for the analysis
increases, 1 iteration takes 3 times as long, 2 iterations 5 times as
long on so forth, since each order of iteration requires one more forward
and backward transform.
The spherical harmonics evaluation uses a
recurrence on associated Legendre polynomials
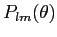 .
This recurrence consumes most of the CPU time used by anafast.
We have therefore included an option to load precomputed values for the .
This recurrence consumes most of the CPU time used by anafast.
We have therefore included an option to load precomputed values for the
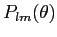 from a file generated by the HEALPix facility
plmgen. This represents a tradeoff between memory usage and execution
speed of anafast. from a file generated by the HEALPix facility
plmgen. This represents a tradeoff between memory usage and execution
speed of anafast.
DATASETS The following datasets are involved in the anafast
processing.
| Dataset |
Description |
|---|
| |
|
|
data/weight_ring_n0xxxx.fits |
Files containing ring weights
for the anafast improved quadrature mode. |
| |
|
|
|
SUPPORT This section lists those routines and facilities (including those external to the Healpix distribution) which can assist in the utilisation of anafast.
-
synfast
- This HEALPix facility can generate a map for analysis by anafast.
-
alteralm
- This HEALPix Fortran facility can be
used to modify the alm extracted by anafast before they are used as
constraints on a synfast run.
-
plmgen
- This HEALPix facility can be used to generate precomputed Legendre polynomials.
EXAMPLE # 1:
Anafast runs in interactive mode -- self-explanatory.
EXAMPLE # 2:
When 'filename' is present, anafast enters the non-interactive mode and parses
its inputs from the file 'filename'. This has the following
structure: the first entry is a qualifier which announces to the parser
which input immediately follows. If this input is omitted in the
input file, the parser assumes the default value.
If the equality sign is omitted, then the parser ignores the entry.
In this way comments may also be included in the file.
In this example, the file contains the following qualifiers:
simul_type = 1
nlmax = 64
theta_cut_deg = 0
iter_order = 0
infile = map.fits
outfile = cl_out.fits
regression = 0
Anafast reads the map from map.fits, makes an analysis and produces CTls up to l=64.
This powerspectrum is saved in the file cl_out.fits.
No galactic cut is excised and no iterations are performed.
As regression is set to 0 (its default value) the map is analyzed as
is, without prior best fit removal of the monopole nor the dipole.
Since
infile2
outfile_alms
outfile_alms2
w8file
w8filedir
plmfile
maskfile
were omitted, they take their default values (empty strings).
This means that no file for precomputed
Legendre polynomials is read, no second map is read, no mask is applied, and anafast does not save the alm values
from the analysis.
Also since
won
is not given, it takes it default value 2, which means that ring
weights are not used.
RELEASE NOTES
 Initial release (HEALPix 0.90)
Initial release (HEALPix 0.90)
 Optional non-interactive operation. Proper FITS file
support. Improved reccurence algorithm for
Optional non-interactive operation. Proper FITS file
support. Improved reccurence algorithm for
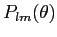 which can compute to higher l values. New functionality: arbitrary order of iterations, precomputed
Plm, dumping of alm. (HEALPix 1.00) which can compute to higher l values. New functionality: arbitrary order of iterations, precomputed
Plm, dumping of alm. (HEALPix 1.00)
 New functionality: possibility of removing the best fit monopole
and dipole. New Parser. Can be linked to FFTW (HEALPix 1.20)
New functionality: possibility of removing the best fit monopole
and dipole. New Parser. Can be linked to FFTW (HEALPix 1.20)
 New functionality: addition of maskfile (HEALPix 2.0)
New functionality: addition of maskfile (HEALPix 2.0)
 Bug correction: correct interaction of iterative scheme with masked pixels (HEALPix 2.01)
Bug correction: correct interaction of iterative scheme with masked pixels (HEALPix 2.01)
 New functionality: cross-correlation of 2 maps; Correction of this documentation: the code expects maskfile and
not mask_file (HEALPix 2.1)
New functionality: cross-correlation of 2 maps; Correction of this documentation: the code expects maskfile and
not mask_file (HEALPix 2.1)
MESSAGESThis section describes error messages generated by anafast
| Message |
Severity |
Text |
|---|
| |
|
|
|
can not allocate memory for array xxx |
Fatal |
You do not have
sufficient system resources to run this
facility at the map resolution you required.
Try a lower map resolution. |
| |
|
|
|
|
|
Eric Hivon
2010-06-18
|

|
Initial release (HEALPix 0.90)
Optional non-interactive operation. Proper FITS file support. Improved reccurence algorithm for
which can compute to higher l values. New functionality: arbitrary order of iterations, precomputed Plm, dumping of alm. (HEALPix 1.00)
New functionality: possibility of removing the best fit monopole and dipole. New Parser. Can be linked to FFTW (HEALPix 1.20)
New functionality: addition of maskfile (HEALPix 2.0)
Bug correction: correct interaction of iterative scheme with masked pixels (HEALPix 2.01)
New functionality: cross-correlation of 2 maps; Correction of this documentation: the code expects maskfile and not mask_file (HEALPix 2.1)